{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Introduction to machine learning with python"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Question: When we talk about machine learning, what method do you think about?"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
" "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Deep learning model\n",
"\n",
"\n",
"More specifically, we will use some classification models to expain the main concepts of deep learning."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Mutilayer Perceptron (MLP)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
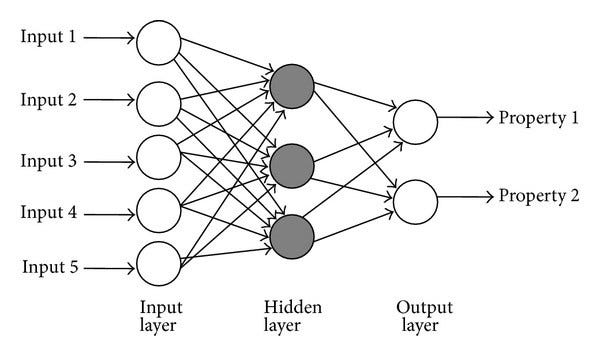
"The most basic model structure is what we call Multilayer Perceptron\n",
"
"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Deep learning model\n",
"\n",
"\n",
"More specifically, we will use some classification models to expain the main concepts of deep learning."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Mutilayer Perceptron (MLP)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The most basic model structure is what we call Multilayer Perceptron\n",
" \n",
"\n",
"Its constructed by basic units that are called 'Neurons' or 'Perceptrons'. Each individual one is actually very simply structured.\n",
"\n",
"
\n",
"\n",
"Its constructed by basic units that are called 'Neurons' or 'Perceptrons'. Each individual one is actually very simply structured.\n",
"\n",
" "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Although each one is a very simple function approximater, by layering them together, the model now has a much greater power and flexibility in approximating functions.\n",
"\n",
"The entire learning process can be divided into three main parts:\n",
"* Forward propagation (Forward pass)\n",
"* Calculation of the loss function\n",
"* Backward propagation (Backward pass/Backpropagation)\n",
"\n",
"
"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Although each one is a very simple function approximater, by layering them together, the model now has a much greater power and flexibility in approximating functions.\n",
"\n",
"The entire learning process can be divided into three main parts:\n",
"* Forward propagation (Forward pass)\n",
"* Calculation of the loss function\n",
"* Backward propagation (Backward pass/Backpropagation)\n",
"\n",
"  \n",
"\n",
"But after all, neural networks can be thing of as a function approximator, but with less assumptions.\n",
"A fun app for visualization MLP: https://playground.tensorflow.org/"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Convolutional Layer"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"MLP isn't the best fit on all problems. Especially things like images. How do we get 'feature' from images? Per pixel? Thats just way too much. So another kind of layer comes into play.\n",
"\n",
"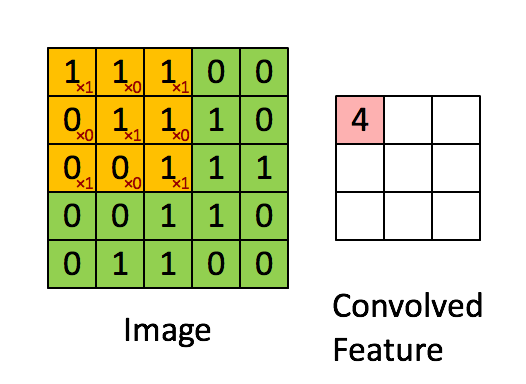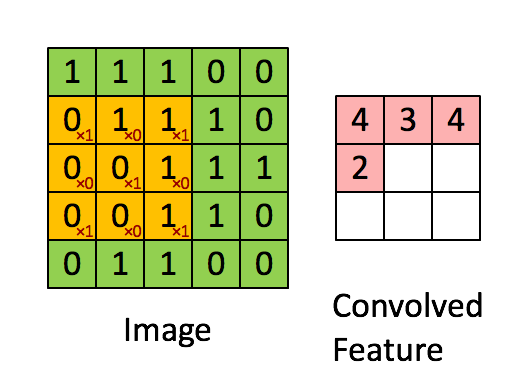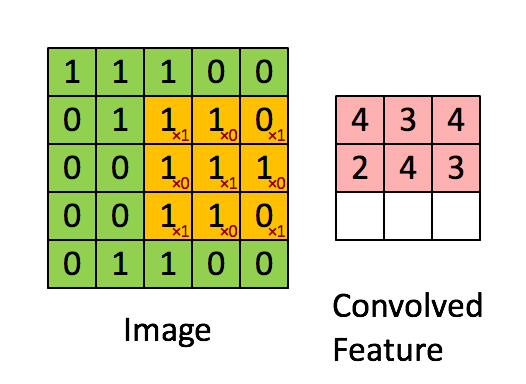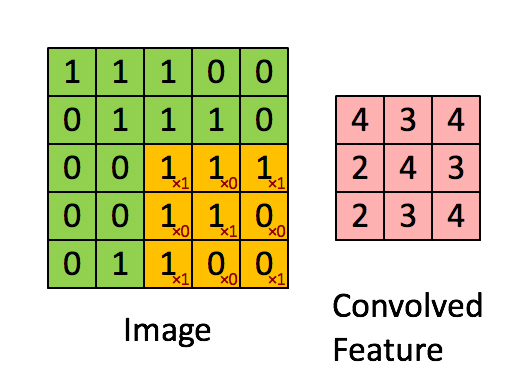\n",
"\n",
"We call the weight we have learnt 'filters'. It to some extent represents what pattern the model has learnt to look for. And there are usually multiple filters per convolution layer, give the model the ability to learn more than one pattern.\n",
"\n",
"
\n",
"\n",
"But after all, neural networks can be thing of as a function approximator, but with less assumptions.\n",
"A fun app for visualization MLP: https://playground.tensorflow.org/"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Convolutional Layer"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"MLP isn't the best fit on all problems. Especially things like images. How do we get 'feature' from images? Per pixel? Thats just way too much. So another kind of layer comes into play.\n",
"\n",
"\n",
"\n",
"We call the weight we have learnt 'filters'. It to some extent represents what pattern the model has learnt to look for. And there are usually multiple filters per convolution layer, give the model the ability to learn more than one pattern.\n",
"\n",
" "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Pooling layer\n",
"\n",
"As seen in the above figure structure, pooling layers often exist in deep learning models. Its main usage is to condense information. There are several ways pooling can be done:\n",
"\n",
"
"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Pooling layer\n",
"\n",
"As seen in the above figure structure, pooling layers often exist in deep learning models. Its main usage is to condense information. There are several ways pooling can be done:\n",
"\n",
" \n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Convolutional Neural Networks for regulatory genomics\n",
"\n",
"In this part, we will walk through how to build, train, and evaluate a convolutional neural network (CNN) for the computational task of predicting transcription factor binding sites from Chromatin Immunoprecipitation sequencing (ChIP-seq) data.\n",
"\n",
"We will employ a CNN model to experimental ChIP-seq data for CTCF, a transciption factor that is known to bind to DNA and help form loops. We will set this up as a supervised learning task, specifically a binary classification, where we give the model a sequence and it has to predict whether or not it is associated with TF binding (i.e. presence or absence of a ChIP-seq peak). Thus, we need a set of background sequences which don't have a ChIP-seq peak. \n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import h5py # python module needed to load dataset (stored in an hdf5 file format)\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"%matplotlib inline"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!wget https://www.dropbox.com/s/0ks4dk6md2l9mqx/CTCF_200.h5"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# load dataset\n",
"file_path = 'CTCF_200.h5'\n",
"dataset = h5py.File(file_path, 'r')\n",
"x_train = np.array(dataset['x_train']).transpose([0,2,1])\n",
"y_train = np.array(dataset['y_train'])\n",
"x_valid = np.array(dataset['x_valid']).transpose([0,2,1])\n",
"y_valid = np.array(dataset['y_valid'])\n",
"x_test = np.array(dataset['x_test']).transpose([0,2,1])\n",
"y_test = np.array(dataset['y_test'])\n",
"\n",
"alphabet = 'ACGT'\n",
"\n",
"# print shape of training data\n",
"print(x_train.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There should be 51,156 training sequences each 200 nts long with an alphabet size of 4. Let's see what a sequence actually looks like:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(x_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This format is called 'one-hot encoding'. Every column in this 2d array represents on letter in the alphabet 'ACTG'. This is how sequence data is usually treated in deep learning problems"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(y_train.shape)\n",
"print(y_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Neural Network implementation\n",
"\n",
"TensorFlow is a widely used deep learning framework. Meaning that all the function/layers that we have mentioned above is already implemented as easy-to-use functions. For this class we will especially depend on TensorFlow.keras.\n",
"\n",
"There are lots of classes/blogs online for tensorflow tutorial. Tensorflow itself also come with a few tutorials:\n",
"https://www.tensorflow.org/tutorials\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"\n",
"# import modules that will make it easy to build and train the linear regression model\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# let's setup a sequential model -- which means that the model will be feed \n",
"# forward (all models in this course will be sequential)\n",
"model = models.Sequential()\n",
"\n",
"# the input layer specifies the shape of the input data to the model. Since our input \n",
"# features, x, are 1-dimensional, we can set this value to 1. All models require\n",
"# some input layer (where you feed in data).\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"\n",
"# a dense connection to 1 unit means that the inputs are fully connected to the \n",
"# output neuron (in this case a single output neuron). Linear is the default \n",
"# activation, but here we explicitly write it.\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"After we build the model, we can print a summary that shows the order of the layers, the shapes and number of parameters:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"model.summary()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Build loss function\n",
"\n",
"For both training and evaluation, we need to define a loss function that measures how closely the model's predictions match the target classes. This is usually a log-likelihood function for supervised learning. The log-likelihood for a binary classification is the binary cross-entropy loss function\n",
"\n",
"Fortunately, Keras makes this super easy as you'll see below.\n",
"\n",
"### Build optimizer\n",
"\n",
"ADAM is the best introductory optimizer for most tasks. It performs an adaptive learning rate based on previous gradients and hence it is much faster to converge and leads to consistently good solutions. \n",
"\n",
"For more info on adam: https://towardsdatascience.com/adam-latest-trends-in-deep-learning-optimization-6be9a291375c"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now that we built the model, we have to set up the loss function and the optimizer.\n",
"# Since it's a linear regression, we will set it to 'mean_squared_error'. \n",
"# we will set the optimizer to stochastic gradient descent 'sgd' (even though this\n",
"# is unnecessary for a linear regression)\n",
"\n",
"loss = keras.losses.BinaryCrossentropy()\n",
"optimizer = keras.optimizers.Adam(learning_rate=0.001)\n",
"model.compile(loss=loss, optimizer=optimizer, metrics=['accuracy'])\n",
"\n",
"# a short cut is: model.compile(loss='binary_crossentropy', optimizer='adam', metrics=[auroc])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Fit the model\n",
"\n",
"We want to train our CNN model with mini-batch stochastic gradient descent. So, we need a way to generate mini-batches of data. Keras handles all of this for you through the setting batch_size. It also lets you select the number of epochs to train. You can also monitor validation performance by adding the validation_data flag. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now let's fit the model. The main inputs are x, y, epochs, which sets the number \n",
"# of times to run through the dataset with SGD. Also, we set verbose to false,\n",
"# because too many outputs are written and it is overwhelming for a linear model.\n",
"# We will turn this flag to true for other datasets later.\n",
"model.fit(x_train, y_train, batch_size=100, epochs=30, validation_data=(x_valid, y_valid))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now while these saliency plots seem to find a motif pattern that resembles the CTCF motif (see: https://www.factorbook.org/tf/human/CTCF/motif/ENCSR000BIE), the saliency maps are really noisy. From my experience, this arises because of overfitting, which leads to spurious saliency maps as well -- here overfitting does not affect classification performance on the test set and is termed benign overfitting.\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Redo analysis\n",
"\n",
"Let's redo this analysis with some regularization added to our CNN model. A very effective regularization is dropout and early stopping. For convolutional layers, let's add a dropout rate of 0.2 and for dense layers let's use a dropout of 0.5. For early stopping, there is a built in \"callback\" in keras. Callbacks are objects taht perform actions during training. The early stopping call back checks whether the model's validation performance hasn't improved for x number of epochs. if the x number of epochs exceeds your patience, then training loop is stopped. See below for example code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# build model\n",
"model = models.Sequential()\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Dropout(0.2)) # <-- add 0.1-0.2 for conv layers\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"model.add(layers.Dropout(0.5)) # <-- add 0.1-0.5 for dense layers\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))\n",
"\n",
"# compile model\n",
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n",
"\n",
"# create a callback for early stopping. Now we can increase the number of epochs \n",
"# as early stopping will automatically stop training earlier\n",
"es_callback = keras.callbacks.EarlyStopping(monitor='loss', patience=5)\n",
"\n",
"# fit model\n",
"model.fit(x_train, y_train, batch_size=128, epochs=100, \n",
" validation_data=(x_valid, y_valid),\n",
" callbacks=es_callback)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Slight performance difference? Thats normal and expected. The initilization and learning process of a deep learning neural network have random factors in it. But with enough data, the performance would always stablize around the same place"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Validating the model \n",
"\n",
"To validate what the network is learning let's calculate a saliency map -- derivative of the output neuron with respect to the inputs. Luckily, tensorflow makes this easy because it calculates gradients using automatic differentiation. We can calculate the gradient with the function model.optimizer.get_gradients. We need to create a keras function to execute this. See code below:\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import tensorflow\n",
"# create an op to calculate gradients of outputs with respect to inputs (i.e. saliency map)\n",
"# Note that we are taking the derivative of the pre-activated output layer (not the output layer)\n",
"# this is because the sigmoid can make the gradients very small when predictions are close to 1 or 0\n",
"def saliency_map(X,model,func = tensorflow.math.reduce_mean):\n",
" \"\"\"fast function to generate saliency maps\"\"\"\n",
" layer_output=model.get_layer(index = -2).output\n",
" intermediate_model=tensorflow.keras.models.Model(inputs=model.input,outputs=layer_output)\n",
" if not tensorflow.is_tensor(X):\n",
" X = tensorflow.Variable(X)\n",
" with tensorflow.GradientTape() as tape:\n",
" tape.watch(X)\n",
" outputs = func(intermediate_model(X))\n",
" return tape.gradient(outputs, X)\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now we should choose which sequences we want to perform saliency analysis on. Let's choose the top 10 test sequences with the highest predictions. To find these, we can first predict the test sequences, sort them in descending order, then slice the top ten."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# To get model predictions, we can simply call predict.\n",
"predictions = model.predict(x_test)\n",
"\n",
"# sort predictions in decending order\n",
"index = np.argsort(predictions[:,0])[::-1]\n",
"\n",
"# get the top num_plot predictions\n",
"num_plots = 10\n",
"X = x_test[index[:num_plots]]\n",
"\n",
"# calculate saliency maps\n",
"saliency_map = saliency_map(X,model)[0]"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's visualize the saliency maps. We can plot this as a heat map:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"plt.figure(figsize=(20,6))\n",
"plt.imshow(saliency_map.numpy().T)\n",
"plt.yticks([0,1,2,3], alphabet)\n",
"plt.colorbar()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This isn't a very nice visualization. Fortunately for you, [Logomaker](https://logomaker.readthedocs.io/en/latest/) (by Tareen and Kinney) can help us visualize saliency maps. We need to first install it on Google Colab."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!pip install logomaker"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I have created a function to visualize the saliency maps. You are welcome to take a look at it. It essentially just converts the saliency map in the format needed by logomaker. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import logomaker \n",
"\n",
"def plot_saliency_map(scores, alphabet, ax=None):\n",
" L,A = scores.shape\n",
" counts_df = pd.DataFrame(data=0.0, columns=list(alphabet), index=list(range(L)))\n",
" for a in range(A):\n",
" for l in range(L):\n",
" counts_df.iloc[l,a] = scores[l,a]\n",
"\n",
" if not ax:\n",
" ax = plt.subplot(1,1,1)\n",
" logomaker.Logo(counts_df, ax=ax)\n",
"\n",
"\n",
"saliency_scores = (saliency_map * X).numpy() # grad * input\n",
"for scores in saliency_scores:\n",
" fig = plt.figure(figsize=(10,1))\n",
" ax = plt.subplot(1,1,1)\n",
" plot_saliency_map(scores, alphabet, ax)"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.0"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Convolutional Neural Networks for regulatory genomics\n",
"\n",
"In this part, we will walk through how to build, train, and evaluate a convolutional neural network (CNN) for the computational task of predicting transcription factor binding sites from Chromatin Immunoprecipitation sequencing (ChIP-seq) data.\n",
"\n",
"We will employ a CNN model to experimental ChIP-seq data for CTCF, a transciption factor that is known to bind to DNA and help form loops. We will set this up as a supervised learning task, specifically a binary classification, where we give the model a sequence and it has to predict whether or not it is associated with TF binding (i.e. presence or absence of a ChIP-seq peak). Thus, we need a set of background sequences which don't have a ChIP-seq peak. \n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import h5py # python module needed to load dataset (stored in an hdf5 file format)\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"%matplotlib inline"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!wget https://www.dropbox.com/s/0ks4dk6md2l9mqx/CTCF_200.h5"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# load dataset\n",
"file_path = 'CTCF_200.h5'\n",
"dataset = h5py.File(file_path, 'r')\n",
"x_train = np.array(dataset['x_train']).transpose([0,2,1])\n",
"y_train = np.array(dataset['y_train'])\n",
"x_valid = np.array(dataset['x_valid']).transpose([0,2,1])\n",
"y_valid = np.array(dataset['y_valid'])\n",
"x_test = np.array(dataset['x_test']).transpose([0,2,1])\n",
"y_test = np.array(dataset['y_test'])\n",
"\n",
"alphabet = 'ACGT'\n",
"\n",
"# print shape of training data\n",
"print(x_train.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There should be 51,156 training sequences each 200 nts long with an alphabet size of 4. Let's see what a sequence actually looks like:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(x_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This format is called 'one-hot encoding'. Every column in this 2d array represents on letter in the alphabet 'ACTG'. This is how sequence data is usually treated in deep learning problems"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(y_train.shape)\n",
"print(y_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Neural Network implementation\n",
"\n",
"TensorFlow is a widely used deep learning framework. Meaning that all the function/layers that we have mentioned above is already implemented as easy-to-use functions. For this class we will especially depend on TensorFlow.keras.\n",
"\n",
"There are lots of classes/blogs online for tensorflow tutorial. Tensorflow itself also come with a few tutorials:\n",
"https://www.tensorflow.org/tutorials\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"\n",
"# import modules that will make it easy to build and train the linear regression model\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# let's setup a sequential model -- which means that the model will be feed \n",
"# forward (all models in this course will be sequential)\n",
"model = models.Sequential()\n",
"\n",
"# the input layer specifies the shape of the input data to the model. Since our input \n",
"# features, x, are 1-dimensional, we can set this value to 1. All models require\n",
"# some input layer (where you feed in data).\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"\n",
"# a dense connection to 1 unit means that the inputs are fully connected to the \n",
"# output neuron (in this case a single output neuron). Linear is the default \n",
"# activation, but here we explicitly write it.\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"After we build the model, we can print a summary that shows the order of the layers, the shapes and number of parameters:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"model.summary()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Build loss function\n",
"\n",
"For both training and evaluation, we need to define a loss function that measures how closely the model's predictions match the target classes. This is usually a log-likelihood function for supervised learning. The log-likelihood for a binary classification is the binary cross-entropy loss function\n",
"\n",
"Fortunately, Keras makes this super easy as you'll see below.\n",
"\n",
"### Build optimizer\n",
"\n",
"ADAM is the best introductory optimizer for most tasks. It performs an adaptive learning rate based on previous gradients and hence it is much faster to converge and leads to consistently good solutions. \n",
"\n",
"For more info on adam: https://towardsdatascience.com/adam-latest-trends-in-deep-learning-optimization-6be9a291375c"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now that we built the model, we have to set up the loss function and the optimizer.\n",
"# Since it's a linear regression, we will set it to 'mean_squared_error'. \n",
"# we will set the optimizer to stochastic gradient descent 'sgd' (even though this\n",
"# is unnecessary for a linear regression)\n",
"\n",
"loss = keras.losses.BinaryCrossentropy()\n",
"optimizer = keras.optimizers.Adam(learning_rate=0.001)\n",
"model.compile(loss=loss, optimizer=optimizer, metrics=['accuracy'])\n",
"\n",
"# a short cut is: model.compile(loss='binary_crossentropy', optimizer='adam', metrics=[auroc])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Fit the model\n",
"\n",
"We want to train our CNN model with mini-batch stochastic gradient descent. So, we need a way to generate mini-batches of data. Keras handles all of this for you through the setting batch_size. It also lets you select the number of epochs to train. You can also monitor validation performance by adding the validation_data flag. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now let's fit the model. The main inputs are x, y, epochs, which sets the number \n",
"# of times to run through the dataset with SGD. Also, we set verbose to false,\n",
"# because too many outputs are written and it is overwhelming for a linear model.\n",
"# We will turn this flag to true for other datasets later.\n",
"model.fit(x_train, y_train, batch_size=100, epochs=30, validation_data=(x_valid, y_valid))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now while these saliency plots seem to find a motif pattern that resembles the CTCF motif (see: https://www.factorbook.org/tf/human/CTCF/motif/ENCSR000BIE), the saliency maps are really noisy. From my experience, this arises because of overfitting, which leads to spurious saliency maps as well -- here overfitting does not affect classification performance on the test set and is termed benign overfitting.\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Redo analysis\n",
"\n",
"Let's redo this analysis with some regularization added to our CNN model. A very effective regularization is dropout and early stopping. For convolutional layers, let's add a dropout rate of 0.2 and for dense layers let's use a dropout of 0.5. For early stopping, there is a built in \"callback\" in keras. Callbacks are objects taht perform actions during training. The early stopping call back checks whether the model's validation performance hasn't improved for x number of epochs. if the x number of epochs exceeds your patience, then training loop is stopped. See below for example code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# build model\n",
"model = models.Sequential()\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Dropout(0.2)) # <-- add 0.1-0.2 for conv layers\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"model.add(layers.Dropout(0.5)) # <-- add 0.1-0.5 for dense layers\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))\n",
"\n",
"# compile model\n",
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n",
"\n",
"# create a callback for early stopping. Now we can increase the number of epochs \n",
"# as early stopping will automatically stop training earlier\n",
"es_callback = keras.callbacks.EarlyStopping(monitor='loss', patience=5)\n",
"\n",
"# fit model\n",
"model.fit(x_train, y_train, batch_size=128, epochs=100, \n",
" validation_data=(x_valid, y_valid),\n",
" callbacks=es_callback)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Slight performance difference? Thats normal and expected. The initilization and learning process of a deep learning neural network have random factors in it. But with enough data, the performance would always stablize around the same place"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Validating the model \n",
"\n",
"To validate what the network is learning let's calculate a saliency map -- derivative of the output neuron with respect to the inputs. Luckily, tensorflow makes this easy because it calculates gradients using automatic differentiation. We can calculate the gradient with the function model.optimizer.get_gradients. We need to create a keras function to execute this. See code below:\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import tensorflow\n",
"# create an op to calculate gradients of outputs with respect to inputs (i.e. saliency map)\n",
"# Note that we are taking the derivative of the pre-activated output layer (not the output layer)\n",
"# this is because the sigmoid can make the gradients very small when predictions are close to 1 or 0\n",
"def saliency_map(X,model,func = tensorflow.math.reduce_mean):\n",
" \"\"\"fast function to generate saliency maps\"\"\"\n",
" layer_output=model.get_layer(index = -2).output\n",
" intermediate_model=tensorflow.keras.models.Model(inputs=model.input,outputs=layer_output)\n",
" if not tensorflow.is_tensor(X):\n",
" X = tensorflow.Variable(X)\n",
" with tensorflow.GradientTape() as tape:\n",
" tape.watch(X)\n",
" outputs = func(intermediate_model(X))\n",
" return tape.gradient(outputs, X)\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now we should choose which sequences we want to perform saliency analysis on. Let's choose the top 10 test sequences with the highest predictions. To find these, we can first predict the test sequences, sort them in descending order, then slice the top ten."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# To get model predictions, we can simply call predict.\n",
"predictions = model.predict(x_test)\n",
"\n",
"# sort predictions in decending order\n",
"index = np.argsort(predictions[:,0])[::-1]\n",
"\n",
"# get the top num_plot predictions\n",
"num_plots = 10\n",
"X = x_test[index[:num_plots]]\n",
"\n",
"# calculate saliency maps\n",
"saliency_map = saliency_map(X,model)[0]"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's visualize the saliency maps. We can plot this as a heat map:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"plt.figure(figsize=(20,6))\n",
"plt.imshow(saliency_map.numpy().T)\n",
"plt.yticks([0,1,2,3], alphabet)\n",
"plt.colorbar()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This isn't a very nice visualization. Fortunately for you, [Logomaker](https://logomaker.readthedocs.io/en/latest/) (by Tareen and Kinney) can help us visualize saliency maps. We need to first install it on Google Colab."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!pip install logomaker"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I have created a function to visualize the saliency maps. You are welcome to take a look at it. It essentially just converts the saliency map in the format needed by logomaker. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import logomaker \n",
"\n",
"def plot_saliency_map(scores, alphabet, ax=None):\n",
" L,A = scores.shape\n",
" counts_df = pd.DataFrame(data=0.0, columns=list(alphabet), index=list(range(L)))\n",
" for a in range(A):\n",
" for l in range(L):\n",
" counts_df.iloc[l,a] = scores[l,a]\n",
"\n",
" if not ax:\n",
" ax = plt.subplot(1,1,1)\n",
" logomaker.Logo(counts_df, ax=ax)\n",
"\n",
"\n",
"saliency_scores = (saliency_map * X).numpy() # grad * input\n",
"for scores in saliency_scores:\n",
" fig = plt.figure(figsize=(10,1))\n",
" ax = plt.subplot(1,1,1)\n",
" plot_saliency_map(scores, alphabet, ax)"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.0"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
 \n",
"\n",
"Its constructed by basic units that are called 'Neurons' or 'Perceptrons'. Each individual one is actually very simply structured.\n",
"\n",
"
\n",
"\n",
"Its constructed by basic units that are called 'Neurons' or 'Perceptrons'. Each individual one is actually very simply structured.\n",
"\n",
" "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Although each one is a very simple function approximater, by layering them together, the model now has a much greater power and flexibility in approximating functions.\n",
"\n",
"The entire learning process can be divided into three main parts:\n",
"* Forward propagation (Forward pass)\n",
"* Calculation of the loss function\n",
"* Backward propagation (Backward pass/Backpropagation)\n",
"\n",
"
"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Although each one is a very simple function approximater, by layering them together, the model now has a much greater power and flexibility in approximating functions.\n",
"\n",
"The entire learning process can be divided into three main parts:\n",
"* Forward propagation (Forward pass)\n",
"* Calculation of the loss function\n",
"* Backward propagation (Backward pass/Backpropagation)\n",
"\n",
"  \n",
"\n",
"But after all, neural networks can be thing of as a function approximator, but with less assumptions.\n",
"A fun app for visualization MLP: https://playground.tensorflow.org/"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Convolutional Layer"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"MLP isn't the best fit on all problems. Especially things like images. How do we get 'feature' from images? Per pixel? Thats just way too much. So another kind of layer comes into play.\n",
"\n",
"\n",
"\n",
"We call the weight we have learnt 'filters'. It to some extent represents what pattern the model has learnt to look for. And there are usually multiple filters per convolution layer, give the model the ability to learn more than one pattern.\n",
"\n",
"
\n",
"\n",
"But after all, neural networks can be thing of as a function approximator, but with less assumptions.\n",
"A fun app for visualization MLP: https://playground.tensorflow.org/"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Convolutional Layer"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"MLP isn't the best fit on all problems. Especially things like images. How do we get 'feature' from images? Per pixel? Thats just way too much. So another kind of layer comes into play.\n",
"\n",
"\n",
"\n",
"We call the weight we have learnt 'filters'. It to some extent represents what pattern the model has learnt to look for. And there are usually multiple filters per convolution layer, give the model the ability to learn more than one pattern.\n",
"\n",
" \n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Convolutional Neural Networks for regulatory genomics\n",
"\n",
"In this part, we will walk through how to build, train, and evaluate a convolutional neural network (CNN) for the computational task of predicting transcription factor binding sites from Chromatin Immunoprecipitation sequencing (ChIP-seq) data.\n",
"\n",
"We will employ a CNN model to experimental ChIP-seq data for CTCF, a transciption factor that is known to bind to DNA and help form loops. We will set this up as a supervised learning task, specifically a binary classification, where we give the model a sequence and it has to predict whether or not it is associated with TF binding (i.e. presence or absence of a ChIP-seq peak). Thus, we need a set of background sequences which don't have a ChIP-seq peak. \n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import h5py # python module needed to load dataset (stored in an hdf5 file format)\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"%matplotlib inline"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!wget https://www.dropbox.com/s/0ks4dk6md2l9mqx/CTCF_200.h5"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# load dataset\n",
"file_path = 'CTCF_200.h5'\n",
"dataset = h5py.File(file_path, 'r')\n",
"x_train = np.array(dataset['x_train']).transpose([0,2,1])\n",
"y_train = np.array(dataset['y_train'])\n",
"x_valid = np.array(dataset['x_valid']).transpose([0,2,1])\n",
"y_valid = np.array(dataset['y_valid'])\n",
"x_test = np.array(dataset['x_test']).transpose([0,2,1])\n",
"y_test = np.array(dataset['y_test'])\n",
"\n",
"alphabet = 'ACGT'\n",
"\n",
"# print shape of training data\n",
"print(x_train.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There should be 51,156 training sequences each 200 nts long with an alphabet size of 4. Let's see what a sequence actually looks like:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(x_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This format is called 'one-hot encoding'. Every column in this 2d array represents on letter in the alphabet 'ACTG'. This is how sequence data is usually treated in deep learning problems"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(y_train.shape)\n",
"print(y_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Neural Network implementation\n",
"\n",
"TensorFlow is a widely used deep learning framework. Meaning that all the function/layers that we have mentioned above is already implemented as easy-to-use functions. For this class we will especially depend on TensorFlow.keras.\n",
"\n",
"There are lots of classes/blogs online for tensorflow tutorial. Tensorflow itself also come with a few tutorials:\n",
"https://www.tensorflow.org/tutorials\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"\n",
"# import modules that will make it easy to build and train the linear regression model\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# let's setup a sequential model -- which means that the model will be feed \n",
"# forward (all models in this course will be sequential)\n",
"model = models.Sequential()\n",
"\n",
"# the input layer specifies the shape of the input data to the model. Since our input \n",
"# features, x, are 1-dimensional, we can set this value to 1. All models require\n",
"# some input layer (where you feed in data).\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"\n",
"# a dense connection to 1 unit means that the inputs are fully connected to the \n",
"# output neuron (in this case a single output neuron). Linear is the default \n",
"# activation, but here we explicitly write it.\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"After we build the model, we can print a summary that shows the order of the layers, the shapes and number of parameters:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"model.summary()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Build loss function\n",
"\n",
"For both training and evaluation, we need to define a loss function that measures how closely the model's predictions match the target classes. This is usually a log-likelihood function for supervised learning. The log-likelihood for a binary classification is the binary cross-entropy loss function\n",
"\n",
"Fortunately, Keras makes this super easy as you'll see below.\n",
"\n",
"### Build optimizer\n",
"\n",
"ADAM is the best introductory optimizer for most tasks. It performs an adaptive learning rate based on previous gradients and hence it is much faster to converge and leads to consistently good solutions. \n",
"\n",
"For more info on adam: https://towardsdatascience.com/adam-latest-trends-in-deep-learning-optimization-6be9a291375c"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now that we built the model, we have to set up the loss function and the optimizer.\n",
"# Since it's a linear regression, we will set it to 'mean_squared_error'. \n",
"# we will set the optimizer to stochastic gradient descent 'sgd' (even though this\n",
"# is unnecessary for a linear regression)\n",
"\n",
"loss = keras.losses.BinaryCrossentropy()\n",
"optimizer = keras.optimizers.Adam(learning_rate=0.001)\n",
"model.compile(loss=loss, optimizer=optimizer, metrics=['accuracy'])\n",
"\n",
"# a short cut is: model.compile(loss='binary_crossentropy', optimizer='adam', metrics=[auroc])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Fit the model\n",
"\n",
"We want to train our CNN model with mini-batch stochastic gradient descent. So, we need a way to generate mini-batches of data. Keras handles all of this for you through the setting batch_size. It also lets you select the number of epochs to train. You can also monitor validation performance by adding the validation_data flag. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now let's fit the model. The main inputs are x, y, epochs, which sets the number \n",
"# of times to run through the dataset with SGD. Also, we set verbose to false,\n",
"# because too many outputs are written and it is overwhelming for a linear model.\n",
"# We will turn this flag to true for other datasets later.\n",
"model.fit(x_train, y_train, batch_size=100, epochs=30, validation_data=(x_valid, y_valid))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now while these saliency plots seem to find a motif pattern that resembles the CTCF motif (see: https://www.factorbook.org/tf/human/CTCF/motif/ENCSR000BIE), the saliency maps are really noisy. From my experience, this arises because of overfitting, which leads to spurious saliency maps as well -- here overfitting does not affect classification performance on the test set and is termed benign overfitting.\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Redo analysis\n",
"\n",
"Let's redo this analysis with some regularization added to our CNN model. A very effective regularization is dropout and early stopping. For convolutional layers, let's add a dropout rate of 0.2 and for dense layers let's use a dropout of 0.5. For early stopping, there is a built in \"callback\" in keras. Callbacks are objects taht perform actions during training. The early stopping call back checks whether the model's validation performance hasn't improved for x number of epochs. if the x number of epochs exceeds your patience, then training loop is stopped. See below for example code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# build model\n",
"model = models.Sequential()\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Dropout(0.2)) # <-- add 0.1-0.2 for conv layers\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"model.add(layers.Dropout(0.5)) # <-- add 0.1-0.5 for dense layers\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))\n",
"\n",
"# compile model\n",
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n",
"\n",
"# create a callback for early stopping. Now we can increase the number of epochs \n",
"# as early stopping will automatically stop training earlier\n",
"es_callback = keras.callbacks.EarlyStopping(monitor='loss', patience=5)\n",
"\n",
"# fit model\n",
"model.fit(x_train, y_train, batch_size=128, epochs=100, \n",
" validation_data=(x_valid, y_valid),\n",
" callbacks=es_callback)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Slight performance difference? Thats normal and expected. The initilization and learning process of a deep learning neural network have random factors in it. But with enough data, the performance would always stablize around the same place"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Validating the model \n",
"\n",
"To validate what the network is learning let's calculate a saliency map -- derivative of the output neuron with respect to the inputs. Luckily, tensorflow makes this easy because it calculates gradients using automatic differentiation. We can calculate the gradient with the function model.optimizer.get_gradients. We need to create a keras function to execute this. See code below:\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import tensorflow\n",
"# create an op to calculate gradients of outputs with respect to inputs (i.e. saliency map)\n",
"# Note that we are taking the derivative of the pre-activated output layer (not the output layer)\n",
"# this is because the sigmoid can make the gradients very small when predictions are close to 1 or 0\n",
"def saliency_map(X,model,func = tensorflow.math.reduce_mean):\n",
" \"\"\"fast function to generate saliency maps\"\"\"\n",
" layer_output=model.get_layer(index = -2).output\n",
" intermediate_model=tensorflow.keras.models.Model(inputs=model.input,outputs=layer_output)\n",
" if not tensorflow.is_tensor(X):\n",
" X = tensorflow.Variable(X)\n",
" with tensorflow.GradientTape() as tape:\n",
" tape.watch(X)\n",
" outputs = func(intermediate_model(X))\n",
" return tape.gradient(outputs, X)\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now we should choose which sequences we want to perform saliency analysis on. Let's choose the top 10 test sequences with the highest predictions. To find these, we can first predict the test sequences, sort them in descending order, then slice the top ten."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# To get model predictions, we can simply call predict.\n",
"predictions = model.predict(x_test)\n",
"\n",
"# sort predictions in decending order\n",
"index = np.argsort(predictions[:,0])[::-1]\n",
"\n",
"# get the top num_plot predictions\n",
"num_plots = 10\n",
"X = x_test[index[:num_plots]]\n",
"\n",
"# calculate saliency maps\n",
"saliency_map = saliency_map(X,model)[0]"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's visualize the saliency maps. We can plot this as a heat map:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"plt.figure(figsize=(20,6))\n",
"plt.imshow(saliency_map.numpy().T)\n",
"plt.yticks([0,1,2,3], alphabet)\n",
"plt.colorbar()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This isn't a very nice visualization. Fortunately for you, [Logomaker](https://logomaker.readthedocs.io/en/latest/) (by Tareen and Kinney) can help us visualize saliency maps. We need to first install it on Google Colab."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!pip install logomaker"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I have created a function to visualize the saliency maps. You are welcome to take a look at it. It essentially just converts the saliency map in the format needed by logomaker. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import logomaker \n",
"\n",
"def plot_saliency_map(scores, alphabet, ax=None):\n",
" L,A = scores.shape\n",
" counts_df = pd.DataFrame(data=0.0, columns=list(alphabet), index=list(range(L)))\n",
" for a in range(A):\n",
" for l in range(L):\n",
" counts_df.iloc[l,a] = scores[l,a]\n",
"\n",
" if not ax:\n",
" ax = plt.subplot(1,1,1)\n",
" logomaker.Logo(counts_df, ax=ax)\n",
"\n",
"\n",
"saliency_scores = (saliency_map * X).numpy() # grad * input\n",
"for scores in saliency_scores:\n",
" fig = plt.figure(figsize=(10,1))\n",
" ax = plt.subplot(1,1,1)\n",
" plot_saliency_map(scores, alphabet, ax)"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.0"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Convolutional Neural Networks for regulatory genomics\n",
"\n",
"In this part, we will walk through how to build, train, and evaluate a convolutional neural network (CNN) for the computational task of predicting transcription factor binding sites from Chromatin Immunoprecipitation sequencing (ChIP-seq) data.\n",
"\n",
"We will employ a CNN model to experimental ChIP-seq data for CTCF, a transciption factor that is known to bind to DNA and help form loops. We will set this up as a supervised learning task, specifically a binary classification, where we give the model a sequence and it has to predict whether or not it is associated with TF binding (i.e. presence or absence of a ChIP-seq peak). Thus, we need a set of background sequences which don't have a ChIP-seq peak. \n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import h5py # python module needed to load dataset (stored in an hdf5 file format)\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"%matplotlib inline"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!wget https://www.dropbox.com/s/0ks4dk6md2l9mqx/CTCF_200.h5"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# load dataset\n",
"file_path = 'CTCF_200.h5'\n",
"dataset = h5py.File(file_path, 'r')\n",
"x_train = np.array(dataset['x_train']).transpose([0,2,1])\n",
"y_train = np.array(dataset['y_train'])\n",
"x_valid = np.array(dataset['x_valid']).transpose([0,2,1])\n",
"y_valid = np.array(dataset['y_valid'])\n",
"x_test = np.array(dataset['x_test']).transpose([0,2,1])\n",
"y_test = np.array(dataset['y_test'])\n",
"\n",
"alphabet = 'ACGT'\n",
"\n",
"# print shape of training data\n",
"print(x_train.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There should be 51,156 training sequences each 200 nts long with an alphabet size of 4. Let's see what a sequence actually looks like:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(x_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This format is called 'one-hot encoding'. Every column in this 2d array represents on letter in the alphabet 'ACTG'. This is how sequence data is usually treated in deep learning problems"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(y_train.shape)\n",
"print(y_train)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Neural Network implementation\n",
"\n",
"TensorFlow is a widely used deep learning framework. Meaning that all the function/layers that we have mentioned above is already implemented as easy-to-use functions. For this class we will especially depend on TensorFlow.keras.\n",
"\n",
"There are lots of classes/blogs online for tensorflow tutorial. Tensorflow itself also come with a few tutorials:\n",
"https://www.tensorflow.org/tutorials\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"\n",
"# import modules that will make it easy to build and train the linear regression model\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# let's setup a sequential model -- which means that the model will be feed \n",
"# forward (all models in this course will be sequential)\n",
"model = models.Sequential()\n",
"\n",
"# the input layer specifies the shape of the input data to the model. Since our input \n",
"# features, x, are 1-dimensional, we can set this value to 1. All models require\n",
"# some input layer (where you feed in data).\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"\n",
"# a dense connection to 1 unit means that the inputs are fully connected to the \n",
"# output neuron (in this case a single output neuron). Linear is the default \n",
"# activation, but here we explicitly write it.\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"After we build the model, we can print a summary that shows the order of the layers, the shapes and number of parameters:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"model.summary()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Build loss function\n",
"\n",
"For both training and evaluation, we need to define a loss function that measures how closely the model's predictions match the target classes. This is usually a log-likelihood function for supervised learning. The log-likelihood for a binary classification is the binary cross-entropy loss function\n",
"\n",
"Fortunately, Keras makes this super easy as you'll see below.\n",
"\n",
"### Build optimizer\n",
"\n",
"ADAM is the best introductory optimizer for most tasks. It performs an adaptive learning rate based on previous gradients and hence it is much faster to converge and leads to consistently good solutions. \n",
"\n",
"For more info on adam: https://towardsdatascience.com/adam-latest-trends-in-deep-learning-optimization-6be9a291375c"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now that we built the model, we have to set up the loss function and the optimizer.\n",
"# Since it's a linear regression, we will set it to 'mean_squared_error'. \n",
"# we will set the optimizer to stochastic gradient descent 'sgd' (even though this\n",
"# is unnecessary for a linear regression)\n",
"\n",
"loss = keras.losses.BinaryCrossentropy()\n",
"optimizer = keras.optimizers.Adam(learning_rate=0.001)\n",
"model.compile(loss=loss, optimizer=optimizer, metrics=['accuracy'])\n",
"\n",
"# a short cut is: model.compile(loss='binary_crossentropy', optimizer='adam', metrics=[auroc])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Fit the model\n",
"\n",
"We want to train our CNN model with mini-batch stochastic gradient descent. So, we need a way to generate mini-batches of data. Keras handles all of this for you through the setting batch_size. It also lets you select the number of epochs to train. You can also monitor validation performance by adding the validation_data flag. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Now let's fit the model. The main inputs are x, y, epochs, which sets the number \n",
"# of times to run through the dataset with SGD. Also, we set verbose to false,\n",
"# because too many outputs are written and it is overwhelming for a linear model.\n",
"# We will turn this flag to true for other datasets later.\n",
"model.fit(x_train, y_train, batch_size=100, epochs=30, validation_data=(x_valid, y_valid))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now while these saliency plots seem to find a motif pattern that resembles the CTCF motif (see: https://www.factorbook.org/tf/human/CTCF/motif/ENCSR000BIE), the saliency maps are really noisy. From my experience, this arises because of overfitting, which leads to spurious saliency maps as well -- here overfitting does not affect classification performance on the test set and is termed benign overfitting.\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Redo analysis\n",
"\n",
"Let's redo this analysis with some regularization added to our CNN model. A very effective regularization is dropout and early stopping. For convolutional layers, let's add a dropout rate of 0.2 and for dense layers let's use a dropout of 0.5. For early stopping, there is a built in \"callback\" in keras. Callbacks are objects taht perform actions during training. The early stopping call back checks whether the model's validation performance hasn't improved for x number of epochs. if the x number of epochs exceeds your patience, then training loop is stopped. See below for example code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from tensorflow import keras\n",
"from tensorflow.keras import models, layers\n",
"\n",
"# build model\n",
"model = models.Sequential()\n",
"model.add(layers.InputLayer(input_shape=(200,4)))\n",
"model.add(layers.Conv1D(filters=24, kernel_size=19, activation='relu', padding='same'))\n",
"model.add(layers.MaxPool1D(pool_size=25))\n",
"model.add(layers.Dropout(0.2)) # <-- add 0.1-0.2 for conv layers\n",
"model.add(layers.Flatten())\n",
"model.add(layers.Dense(units=64, activation='relu'))\n",
"model.add(layers.Dropout(0.5)) # <-- add 0.1-0.5 for dense layers\n",
"model.add(layers.Dense(units=1, activation='linear'))\n",
"model.add(layers.Activation('sigmoid'))\n",
"\n",
"# compile model\n",
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n",
"\n",
"# create a callback for early stopping. Now we can increase the number of epochs \n",
"# as early stopping will automatically stop training earlier\n",
"es_callback = keras.callbacks.EarlyStopping(monitor='loss', patience=5)\n",
"\n",
"# fit model\n",
"model.fit(x_train, y_train, batch_size=128, epochs=100, \n",
" validation_data=(x_valid, y_valid),\n",
" callbacks=es_callback)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# evaluate model\n",
"model.evaluate(x_test, y_test)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Slight performance difference? Thats normal and expected. The initilization and learning process of a deep learning neural network have random factors in it. But with enough data, the performance would always stablize around the same place"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Validating the model \n",
"\n",
"To validate what the network is learning let's calculate a saliency map -- derivative of the output neuron with respect to the inputs. Luckily, tensorflow makes this easy because it calculates gradients using automatic differentiation. We can calculate the gradient with the function model.optimizer.get_gradients. We need to create a keras function to execute this. See code below:\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import tensorflow\n",
"# create an op to calculate gradients of outputs with respect to inputs (i.e. saliency map)\n",
"# Note that we are taking the derivative of the pre-activated output layer (not the output layer)\n",
"# this is because the sigmoid can make the gradients very small when predictions are close to 1 or 0\n",
"def saliency_map(X,model,func = tensorflow.math.reduce_mean):\n",
" \"\"\"fast function to generate saliency maps\"\"\"\n",
" layer_output=model.get_layer(index = -2).output\n",
" intermediate_model=tensorflow.keras.models.Model(inputs=model.input,outputs=layer_output)\n",
" if not tensorflow.is_tensor(X):\n",
" X = tensorflow.Variable(X)\n",
" with tensorflow.GradientTape() as tape:\n",
" tape.watch(X)\n",
" outputs = func(intermediate_model(X))\n",
" return tape.gradient(outputs, X)\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now we should choose which sequences we want to perform saliency analysis on. Let's choose the top 10 test sequences with the highest predictions. To find these, we can first predict the test sequences, sort them in descending order, then slice the top ten."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# To get model predictions, we can simply call predict.\n",
"predictions = model.predict(x_test)\n",
"\n",
"# sort predictions in decending order\n",
"index = np.argsort(predictions[:,0])[::-1]\n",
"\n",
"# get the top num_plot predictions\n",
"num_plots = 10\n",
"X = x_test[index[:num_plots]]\n",
"\n",
"# calculate saliency maps\n",
"saliency_map = saliency_map(X,model)[0]"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's visualize the saliency maps. We can plot this as a heat map:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"plt.figure(figsize=(20,6))\n",
"plt.imshow(saliency_map.numpy().T)\n",
"plt.yticks([0,1,2,3], alphabet)\n",
"plt.colorbar()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This isn't a very nice visualization. Fortunately for you, [Logomaker](https://logomaker.readthedocs.io/en/latest/) (by Tareen and Kinney) can help us visualize saliency maps. We need to first install it on Google Colab."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"!pip install logomaker"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I have created a function to visualize the saliency maps. You are welcome to take a look at it. It essentially just converts the saliency map in the format needed by logomaker. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import logomaker \n",
"\n",
"def plot_saliency_map(scores, alphabet, ax=None):\n",
" L,A = scores.shape\n",
" counts_df = pd.DataFrame(data=0.0, columns=list(alphabet), index=list(range(L)))\n",
" for a in range(A):\n",
" for l in range(L):\n",
" counts_df.iloc[l,a] = scores[l,a]\n",
"\n",
" if not ax:\n",
" ax = plt.subplot(1,1,1)\n",
" logomaker.Logo(counts_df, ax=ax)\n",
"\n",
"\n",
"saliency_scores = (saliency_map * X).numpy() # grad * input\n",
"for scores in saliency_scores:\n",
" fig = plt.figure(figsize=(10,1))\n",
" ax = plt.subplot(1,1,1)\n",
" plot_saliency_map(scores, alphabet, ax)"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.0"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
 "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Deep learning model\n",
"
"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Deep learning model\n",
"